PATH-SAFE: Tracking Foodborne Pathogens and Antimicrobial Resistance Microbes
On this page
Skip the menu of subheadings on this page.Committee action required
Paper is for information only. No action required.
Background
Foodborne disease (FBD) is a major public health risk with 2.4 million individual illnesses and more than 16,000 hospitalisations per year. The vast majority of human disease is caused by a handful of pathogens which, in most cases, enter the food chain from farmed animals or the environment. In addition to FBD, the agri-food supply chain also poses a risk for the transmission of antimicrobial resistance (AMR) as it is transmitted through food, animals, humans, or water. The ability to detect and identify pathogens early and to accurately trace FBD outbreaks to their source are critical steps to improve public health and reduce the economic costs associated with them.
AMR presents a serious threat to the health and welfare of both humans and animals and needs to be tackled with an immediate and appropriately robust response. Whilst the UK has made progress in reducing its use of antibiotics in humans and significantly in animals in the last five years, drug-resistant bloodstream infections in humans have increased by 32% from 2015 to 2019. The rise and spread of AMR is creating a new generation of ‘superbugs’ that cannot be treated with existing medicines. The threat is immediate and the impacts of leaving AMR unchecked are wide-ranging and extremely costly, particularly in terms of global health, food sustainability and security, environmental wellbeing, and socio-economic development.
For these reasons, various government departments already undertake surveillance activities (i.e., by taking and analysing samples from food, livestock, and humans) to identify the pathogens causing an illness, to assess levels of contamination or trace the source and transmission pathways of FBD pathogens and AMR. These activities are critical to effecting better control strategies, but recent advances in technology and data management offer the opportunity to create a step change in surveillance, to protect public health.
The Pathogen Surveillance in Agriculture, Food and Environment (PATH-SAFE) programme is a £19.2m Shared Outcomes Fund (SOF) research programme which aims to develop a national surveillance network, using the latest DNA-sequencing technology and environmental sampling to improve the detection, and tracking of foodborne human pathogens and AMR through the whole agri-food system from farm-to-fork. The heart of this 'virtual network will be a new data platform that will permit the analysis, storage and sharing of pathogen sequence and source data, collected from multiple locations across the UK by diverse government and public organisations (incl. FSA, FSS, DHSC, Defra and others across the devolved administrations). This single, user-friendly data system will enable rapid identification and tracking of foodborne pathogens and AMR, improving public health, and minimising the economic and environment impact of outbreaks.
Programme structure:
The lead organisation for the programme is the FSA but the programme will also bring together expertise from Food Standards Scotland (FSS), the Department of Environment, Food and Rural Affairs (Defra), the Department of Health and Social Care (DHSC), UK Health Security Agency (UKHSA), the Environment Agency (EA), the Veterinary Medicines Directorate (VMD) and others. The governance of the programme will include a Delivery Board for operational oversight, a Strategic Board for strategic oversight and approvals and two advisory groups. The programme will also benefit from two fellowships to sit within the programme team, focussing on data and science.
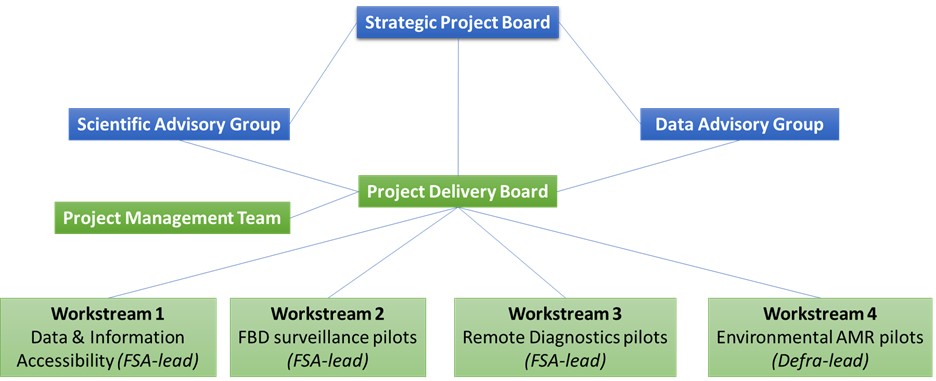
Workstreams:
The programme is made up of four workstreams
|
WS |
Activity |
Lead partners |
Lead fellow |
|
WS1 |
Establish a curated, national foodborne disease genomic data platform |
FSA/FSS |
Data |
|
WS2 |
Develop a pilot infrastructure for regular, multi-location sampling |
FSA/Defra |
Science |
|
WS3 |
Test the feasibility of using portable diagnostics as inspection tools |
FSA/FSS |
Science |
|
WS4 |
Develop a pilot environmental AMR Surveillance system |
Defra/UKHSA/ VMD/EA |
Data and Science to support for this WS but not lead |
Workstream 1: Establish a curated, national foodborne disease genomic data platform
The UK is recognised global leader in genomic database systems. We will utilise this existing expertise, working with academic colleagues and major ‘big data’ stakeholders to create a ‘user-friendly’ platform for the rapid interrogation and archiving of genomic data. We will build on ‘dashboard’ approaches (as developed for COVID-19 monitoring) to create powerful, but easily understood, interfaces that can be used by decision makers (e.g., food inspectors or healthcare professionals). A key element of the data platform development will be allowing the seamlessly integration of sample data with other existing data sources (e.g., infection data, meteorological data etc) to create new knowledge. Mining of pathogen WGS and associated data from existing sources, as well as new sources generated in objective 2, will also be conducted to help populate the new database and enhance data granularity.
Status: Tender document in preparation for bids to develop the data platform, intended to go live next month for work to begin ideally in April. Cross government workshops will be run in March to map user need across the organisation as well as identifying existing data sets for integration with the platform.
Workstream 2: Develop a pilot infrastructure for regular, multi-location sampling
The programme will develop a pilot infrastructure to provide high granularity WGS data from regular, multi-location sampling of wastewater (at primary production sites and environmental water sources); and food products. The workstream will map pathogen populations over time and help refine predictive models that can allow for proactive control. This work will build on existing networks and infrastructure in each of the four nations, such as that already in place for water sampling, including recent UK-wide COVID-19 testing initiatives. Using these existing networks, alongside ongoing food sampling programmes, we will conduct extensive sampling for foodborne pathogens on a regular basis, processed via existing public sector or commercial laboratories, with established sequencing capabilities. Selected samples will be independently validated using standard microbiological testing by FSA/FSS.
Status: Objective setting has taken place with lead partners and workshops will be held across further delivery partners in the coming weeks to assist in developing a specification of work to be undertaken from April 2022.
Workstream 3: Test the feasibility of using portable diagnostics as inspection tools
The programme will investigate the technology readiness levels (TRL) of new portable diagnostics, as well as developing and validating tests for deployment at borders or other remote locations, that would permit rapid testing (results in minutes). This will be achieved by running open competitions across the four nations Utilising existing testing platforms and working with leading academic groups, we will develop fit-for-purpose tests that are ready for field deployment (e.g., at major ports). The co-design of applications with end-users (e.g., policy teams/inspectorates) will be critical to ensure real-world applicability.
Status: The first competition (TRL study) is expected to go live in April/May for work to begin in the summer.
Workstream 4: Develop a pilot environmental AMR Surveillance system
Our overall aim is to create a scientific and evidence-based understanding of the nature and extent of AMR in the environment by 2024 and the drivers that influence this . This pilot will contribute to this aim by delivering an agreed and tested methodology for environmental AMR surveillance, as well as an environmental IT platform that will enable a scaled-up surveillance programme to be undertaken. The pilot intends to obtain new monitoring data on the types and prevalence of AMR in the environment and an IT platform which will house this environmental data. This IT platform will be designed and developed so that it will have the capability to integrate AMR surveillance data collected from humans and animals so that the ambition of having a UK One Health surveillance system for AMR can be realised.
Status: Catchment areas have been selected for the studies and work is underway with the University of Worcester to review work to determine sampling strategy and assessment options for environmental AMR in airborne microorganisms. UKHSA have tendered for the delivery of the One Health platform and bids are under review.
Key outcomes and benefits:
- The programme will provide better data to identify the prevalence, source and pathways of FBD and AMR, helping to prevent spread by enhanced targeting of interventions (e.g., inspections of food businesses, product recalls etc). Other benefits include:
- Improved evidence for informed policy development to enable policy makers to make informed decisions regarding any intervention to prevent or lower risk to human health, as well as improved targeting of regulations through understanding what substances can drive AMR and at what concentrations.
- Reduction in commercial losses, by avoiding inaccurate diagnoses of outbreaks and the associated costs of products recalls and export losses. The pilot project also aims to introduce more rapid testing which can reduce costs for UK importers, as consignments get released faster;
- Strengthening UK Science Excellence, by establishing partnerships with UK businesses to develop new testing tools and by strengthening our reputation as a globally recognised scientific leader;
- Efficient use of resources, by establishing a joint database, so that the same data can be used by multiple departments for different reasons;
- Wider societal benefits, as less FBD cases strengthen the reputation of the UK’s food sector and help secure our £23bn export markets.
- Fewer cases of resistant infections in humans and animals, reducing cost of treatments improved health outcomes and/or epidemic control.
-
Improved agricultural practice including potentially reduced costs both for farmers and water companies through better understanding of concentrations of antimicrobials and pesticides needed to support agriculture without overuse causing pollution and need for additional drinking water treatment. January 2022